Utilizing PGSTE diffusion measurements for molecular weight determination by benchtop NMR
In a previous application note, we discussed how the molecular weight of two PDMS polymers can be measured via end-group analysis in 1D ²⁹Si-NMR. Building on that discussion, this study focuses on the use of diffusion NMR spectroscopy to determine the molecular weight of selected PDMS polymer samples.
In materials science, the molecular weight of polymers is a key property, as it correlates with various physical characteristics. Our previous blog posts have demonstrated that polymer molecular weight can be determined using benchtop NMR through two main approaches: (1) end-group analysis and (2) measurement of the self-diffusion coefficient.
End-group analysis can be carried out using straightforward 1D NMR experiments with ¹H, ¹⁹F, ²⁹Si, or other nuclei. However, in the case of PDMS type polymers the end-group signals in 1D 1H spectra are obscured due to overlap or in 1D 29Si spectra are too weak to detect especially for high molecular weight polymers.
Polydimethylsiloxane (PDMS) is a widely used silicone-based polymer known for its exceptional flexibility, chemical stability, and biocompatibility. Composed of repeating –Si(CH₃)₂–O– units (Scheme 1), PDMS exhibits unique properties such as low glass transition temperature, high thermal stability, and excellent hydrophobicity. These characteristics make it suitable for a broad range of applications, including medical devices, microfluidics, coatings and sealants.
Scheme 1. General molecular structure of dimethylpolysiloxanes.
The molecular weight of Polydimethylsiloxanes critically influences their physical and mechanical properties. As molecular weight increases, PDMS exhibits significantly higher viscosity, enhanced tensile strength, and improved thermal stability due to greater chain entanglement and reduced chain-end effects. Low MW PDMS behaves as a fluid with excellent surface migration and is suited for applications like lubricants and coatings, while high MW PDMS forms elastic, rubber-like materials ideal for sealants and elastomers. Understanding these relationships is essential for optimizing PDMS performance across diverse applications.
Experimental Setup
We would like to thank our collaborators at Shin-Etsu Silicones Europe B.V., Germany, for providing us with ten different PDMS samples, varying in viscosity from 50 cSt to 1,000,000 cSt.
For
the NMR measurements, 5 mg of each sample was dissolved in CDCl₃ at a concentration of 5
mg/mL. The ¹H PGSTE spectra were recorded using the following parameters:
Number of scans: 2, Acquisition time: 3.2 s, Repetition time: 7 s, Number of
gradient steps: 8.
This
resulted in a total measurement time of 2 minutes and 7 seconds per sample. All
measurements were performed with ¹³C decoupling, and the diffusion-specific
parameters were adjusted according to each sample.
NMR Results & Discussion
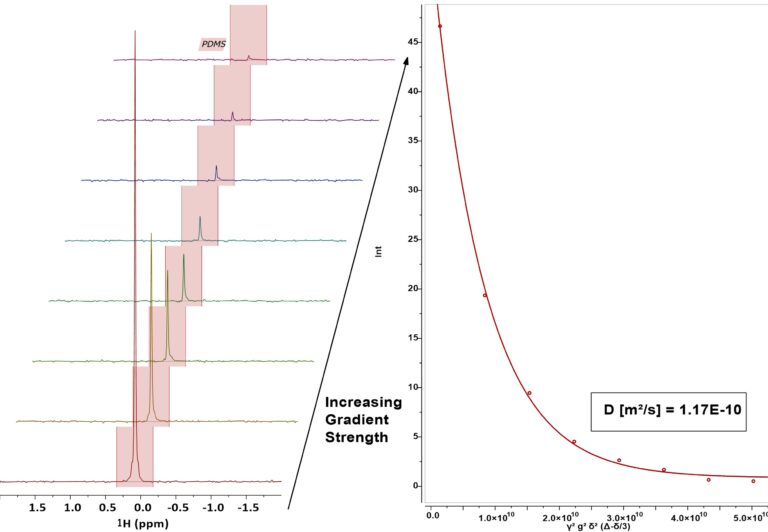
Figure 1. Left: Example 8 steps PGSTE experiment for the 500 cSt PDMS (5 mg/mL in CDCl3) sample. Right: Stejskal-Tanner plot, depicting the signal integral as a function of γ2g2δ2(Δ-δ/3). The data was fitted to obtain the diffusion coefficient (D).
The ten PDMS samples were measured using a Spinsolve 80 MHz Ultra Multi-X Diffusion system. With measurement times of approximately 2 minutes per sample, the system enabled rapid analysis of a large number of samples—even at low concentrations of 5 mg/mL.
Figure 1 exemplarily shows the 1H-PGSTE measurement of the PDMS sample with a viscosity of 500 cSt on the left. A clear stepwise attenuation of the signal can be observed as the applied gradient strength increases. This demonstrates the outstanding sensitivity of the Spinsolve system, which is capable of detecting even strongly attenuated signals with just two scans.
The right side of Figure 1 displays the corresponding Stejskal-Tanner plot for this measurement, including the fitted curve used to determine the diffusion coefficient D, which was calculated to be 1.17 × 10⁻¹⁰ m²/s.
The molecular weight (M) can be determined by plotting log(D) against log(M), following the linear relationship shown in Scheme 2, characterized by a slope (ν) and a y-axis intercept (log(b’)). This type of plot is traditionally obtained by measuring polymer samples of known molecular weight, used as external calibrants at a fixed concentration and temperature.
Recent studies by the groups of Prof. Junkers [1] and Prof. Haddleton [2] go beyond this traditional approach and have demonstrated the use of universal calibration curves that are independent of polymer type, NMR instrument, or operating frequency. This significantly simplifies the determination of molecular weight via PGSTE diffusion NMR, making it more straightforward and broadly applicable across a range of systems.
Scheme 2. Linear behavior between molecular weight M and diffusion coefficient D from the simplified Stokes-Einstein equation.
Table 1 summarizes the results of the PGSTE diffusion measurements. The diffusion coefficients (D) show a variation of over one order of magnitude, ranging from 3.00 × 10⁻¹⁰ to 2.56 × 10⁻¹¹ m²/s for PDMS samples with viscosities between 50 and 1,000,000 cSt. The corresponding molecular weight values were calculated using the universal calibration curve approach implemented in MaDDOSY, developed by the group of Prof. Haddleton at Warwick University [2]. In line with the diffusion coefficients, molecular weights spanning a broad range—from 2,865 to 177,045 g/mol—were determined.
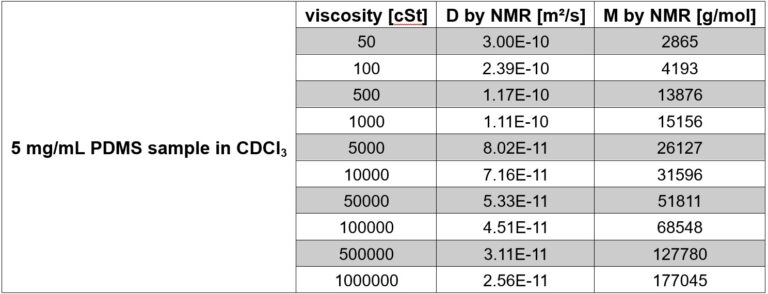
Table 1. Measured diffusion coefficients D and calculated molecular weight values following the universal calibration curve approach from the literature [2].
Figure 2 compares the molecular weight values obtained by NMR with SEC data provided by our collaborators at Shin-Etsu Silicones Europe B.V., Germany. The results clearly demonstrate that the data measured with our Spinsolve Benchtop NMR systems is in excellent agreement with the SEC data across the entire range of tested polymer sizes and viscosity levels.
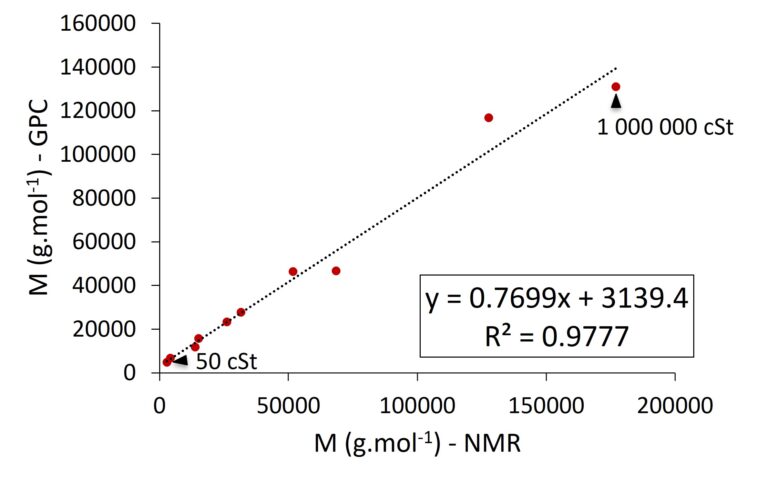
Figure 2. Correlation between the molecular weight values determined by SEC and a Spinsolve 80 MHz Ultra system.
Conclusion
This study demonstrates that diffusion NMR spectroscopy is a robust and non-invasive technique for determining the molecular weight of polydimethylsiloxane (PDMS) polymers. By analyzing diffusion coefficients, it is possible to distinguish between PDMS samples of varying molecular weights and viscosities without the need for chemical derivatization or external calibration. The method provides a valuable complement to traditional techniques such as end-group analysis in ¹D ²⁹Si NMR, particularly for polymers with higher molecular weights or complex structural features where end-group signals may be weak or unresolved.
In addition to its analytical strengths, diffusion NMR offers significant practical advantages. Each measurement takes only two minutes, enabling rapid and efficient analysis. Furthermore, sample preparation is minimal, requiring just a single dilution step. This stands in contrast to techniques like size exclusion chromatography (SEC), which typically involve long measurement times, system equilibration, and external calibration standards, making them more time-consuming and operationally complex.
References
- Solvent-Independent Molecular Weight Determination of Polymers Based on a Truly Universal Calibration; P.-J. Voorter, A. McKay, J. Dai, O. Paravagna, N. R. Cameron, T. Junkers; Angew. Chem. Int. Ed.; Volume 134, Issue 5 (2022) DOI: 10.1002/ange.202114536.
- MaDDOSY (Mass Determination Diffusion Ordered Spectroscopy) using an 80 MHz Bench Top NMR for the Rapid Determination of Polymer, Macromolecular Molecular Weight; O. Tooley, W. Pointer, R. Radmall, M. Hall, V. Beyer, K. Stakem, T. Swift, J. Town, T. Junkers, P. Wilson, D. Lester, D. Haddleton, (2024) DOI: 10.1002/marc.202300692.